Researchers at the University of Liverpool have captured a clear view of the generation process of ‘protein machinery’ that plays a key role in the colonisation of pathogenic Salmonella bacteria.
The findings, published in Nature Communications, answer an important question about how various proteins self-assemble to create a higher-ordered functional organelle in Salmonella to boost metabolism.
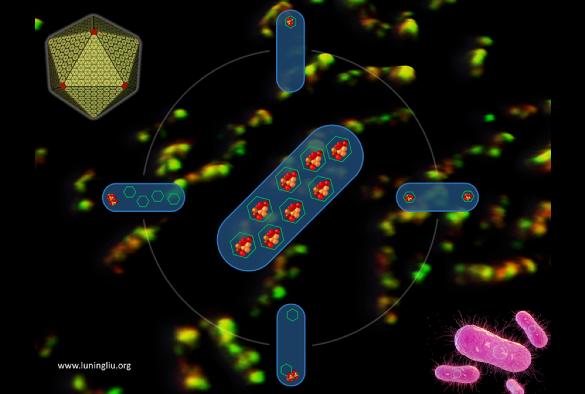
Many pathogenic bacteria, such as Salmonella, use specialised nano-sized organelles, or bacterial microcompartments (BMC). The BMC has a virus-like polyhedral shell made of proteins to encase multiple metabolic cargo enzymes. The protein shell provides a selectively permeable barrier which controls the passage of metabolites and sequesters the reactions in its interior. This ensures higher efficiency of the encapsulated reactions and prevents toxic products from being released into the rest of the cell, providing the pathogens a competitive advantage in human gut.
The research team, led by Professor Luning Liu from the University’s Institute of Systems, Molecular and Integrative Biology, used live-cell fluorescence microscopy and molecular genetics to uncover the de novo stepwise biosynthesis process of propanediol-utilization (PDU) microcompartments, a class of BMCs that are responsible for metabolizing propanediol as a carbon source and recycling of vitamin B12.
“Following our recent work on deciphering the protein stoichiometric composition of PDU microcompartments (Nature Communications 2020, 11:1976), this systematic study provided direct experimental evidence to outline how this microcompartment is formed in Salmonella,” said Professor Luning Liu. “We showed for the first time that the PDU microcompartment undergoes a combination of ‘Shell first’ and ‘Cargo first’ assembly, and identified the key proteins that govern the ordered assembly and encapsulation of cargo enzymes.”
The first author, Dr Mengru Yang, explained: “Salmonella is a dangerous pathogen, and targeting BMC-mediated metabolism will be a promising therapeutic or prophylactic strategy that has yet to be realised. A detailed mechanistic understanding of BMCs is thus required.”
Understanding how the natural microcompartment is generated and modulated in cells could lead to new ways to manipulate BMC biosynthesis in pathogenic bacteria, which could impact the development of novel therapeutic strategies to disrupt metabolism in pathogens.
The research was in collaboration with Professor Jay Hinton at the Institute of Infection, Veterinary and Ecological Sciences, the Centre for Cell Imaging and the Biomedical Electron Microscopy Unit at the Faculty of Health and Life Sciences. The project was funded by Biotechnology and Biological Sciences Research Council (BBSRC), the Royal Society, and Wellcome Trust.
Research reference
“Biogenesis of a bacterial metabolosome for propanediol utilization” is published in Nature Communications 2022, DOI: 10.1038/s41467-022-30608-w