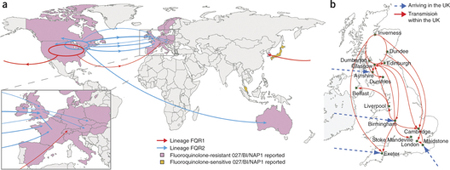
A map tracking the spread of a bacterial strain of clostridium difficile
Scientists at the University of Liverpool have identified the origin of an epidemic strain of Clostridium difficile (027) and its mode of spread using tagging genetic signatures in the DNA of bacteria, some of them associated with its increased virulence.
Clostridium difficile is the most common cause of antibiotic-associated diarrhoea and healthcare-related infection in the developed world. On average, ill patients display symptoms for more than 10 days and a third develop severe disease, including bloody diarrhoea and septicaemia. Until now, however, the key moments in the infection’s evolutionary history have remained unknown.
Outbreaks in North America
Scientists at Liverpool, in collaboration with the Wellcome Trust Sanger Institute, Hinxton, and a number of investigators worldwide, have discovered that a more virulent strain of the bacteria originated from two outbreaks in North America in 2003-4, thought to have been driven by widespread prescription of broad spectrum antibiotics.
This particular strain quickly spread throughout Europe, including a notorious outbreak at Stoke Mandeville Hospital. Using next generation microbial sequencing, evolutionary analysis and epidemiological information, the team identified subtle signatures in the bacteria’s genome that allowed them to pinpoint its worldwide spread.
The data suggests that there were at least four routes of entry for this epidemic strain into UK, three of which arrived independently from North America in Exeter, Ayrshire and Birmingham, and another one which arrived in Maidstone from continental Europe.
Dr Fabio Miyajima, from the University’s Institute of Translational Medicine, said: “We know these organisms are resistant to commonly prescribed antibiotics and disinfectants, including alcohol gel, but perhaps more intriguing is the fact that its rapid spread throughout the world has been more about the ease and frequency with which we travel today.”
Clostridium difficile is air borne and can be transmitted from infected patients to others. Since it survives in the environment for weeks, there is also a strong possibility that healthy individuals who shared a contaminated ambient could transport the spores through clothing, or even on their hands and hair.
He added: “It is important for us to know the ways in which the bacteria can enter various countries so that we can predict just how widespread the infection may become. Importantly, the same techniques can now also be used to track an outbreak within a hospital and community to ensure that it is controlled as rapidly as possible.”
£2.4m grant
Professor Munir Pirmohamed, NHS Chair in Pharmacogenetics and Head of the Wolfson Centre for Personalised Medicine, added: “This is the result of work that was carried out as part of the Liverpool NIHR Biomedical Research Centre. It has now resulted in the award of a £2.4 million program grant from the Medical Research Council, in partnership with the London School of Hygiene and Tropical Medicine and the Wellcome Trust Sanger Institute, which will allow us to build on this work and understand the interactions between the microbial genome, its environment and the patient.
“Our goal is to answer key medical questions, such as why we can’t predict who will contract the condition and why, despite rigorous cleaning procedures to reduce Clostridium difficile, a significant number of people still get it.”
The research is published in Nature Genetics and funded by the Medical Research Council (MRC).