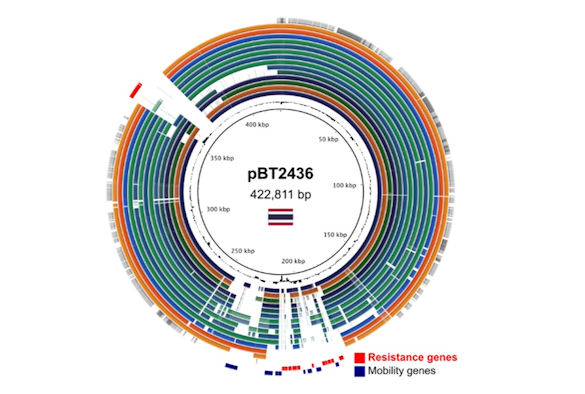
Genome comparison of megaplasmids of the pBT2436-like family
Researchers at the University of Liverpool have gained new insight into how antibiotic resistance evolves and spreads in one of the world’s most dangerous superbugs.
Pseudomonas aeruginosa is one of three top priority pathogens that the World Health Organization deems as “critical” for new antibiotics to target. This difficult-to-treat opportunistic pathogen has the ability to quickly develop resistance to multiple drugs, including powerful ‘last-resort’ carbapenem antibiotics, and poses a great threat to human health.
Alongside the development of new drugs, it is crucial for scientists to gain a better understanding of the factors driving the evolution and spread of antibiotic resistance. A new genomics study published in Nature Communications today has identified a family of plasmids (small circular trands of DNA that replicate independently of chromosomes and can be transferred to other bacteria) responsible for spreading multidrug resistance in P.aeruginosa on a global scale since as far back as the 1970s.
Using a ‘third-generation’ genome sequencing technique called long-read sequencing, the researchers were able to characterise an emergent family of ‘megaplasmids’ contributing to the spread of multi drug resistance in P. aeruginosa in a hospital in Thailand.
Carbapenem-resistant P. aeruginosa is an increasing problem in hospital settings, which provide excellent opportunities for the spread of resistance. Although previous studies have allowed us to study the genes responsible for carbapenem resistance amongst P. aeruginosa clinical isolates, because of the limitations of sequencing technology, we understand less about how the genes spread, and particularly the role of plasmids.
Using long-read sequence data the researchers obtained complete sequences of two closely related megaplasmids (>420 kb) carrying large arrays of antibiotic resistance genes with evidence of extensive duplication and recombination events.
Analysis of DNA sequences deposited in databases reveals the megaplasmid family to be present across clinical, environmental and geographical sources, and in multiple species hosts. The history of the megaplasmid family, inferred from analysis of the available database, suggests that members carrying multiple resistance genes date back to at least the 1970s.
Professor Craig Winstanley, who led the research at the University of Liverpool, said: “Antimicrobial resistance is a huge global issue. If we are to introduce interventions to try to control it, then it is crucial that we understand how the resistance evolves and spreads through bacterial populations. Our study demonstrates the importance of using the best high-resolution sequencing and data analysis procedures to reveal secrets previously hidden amongst the huge amount of data in existing databases.”
Dr Adrian Cazares, who carried out much of the data analysis and co-led the study said: “Bacteria are incredibly adaptable organisms that will use the help of any available tool to thrive. Our findings showcase how bacteria can find these tools in nature, in the form of plasmids, which allow them to collect multiple resistance genes and invade clinical settings causing life-threatening infections. Discovering and targeting these links between the environment and clinic is therefore pivotal for designing better antimicrobial interventions.”
The study involved a collaboration between researchers in Liverpool, Quebec, Canada and Bangkok, Thailand.
This work was supported by the International Pseudomonas Genomics Consortium, funded by Cystic Fibrosis Canada; the Secretaría de Educación, Ciencia, Tecnología e Innovación (SECTEI), Mexico; and the Medical Research Foundation.
The full paper, entitled ‘A megaplasmid family driving dissemination of multidrug resistance in Pseudomonas’, can be found here.