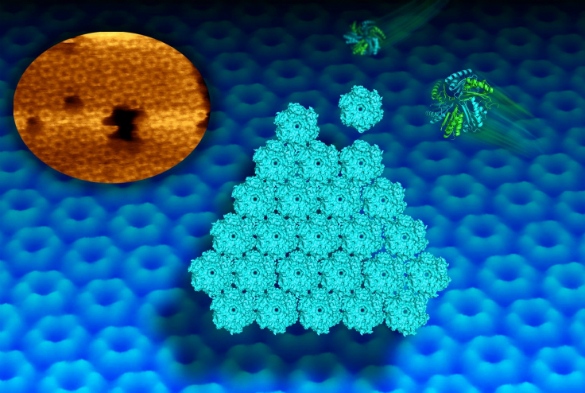
Scientists at the University of Liverpool have used state-of-the-art microscopy to capture the dynamic self-assembly of proteins from tiny bacterial structures, known as bacterial microcompartments (BMCs), which play an important role in metabolism.
The research, which features as the cover article in this week’s Nano Letters, opens up a whole new way of exploring inner-cell architecture and functionality, and could help revolutionise the future of protein-based nanomaterial design and drug delivery in human medicine.
BMCs are widespread among bacterial species and have an outer shell made completely of proteins. Early studies have revealed the structures of individual compartment protein modules, but it is unknown how they self-assemble to build the whole active structure.
Now, a new imaging technique called high-speed atomic force microscopy (AFM) has allowed scientists to picture and video the dynamic building process of the shell facets of the compartments at molecular resolution.
Honeycomb pattern
Matthew Faulkner, a PhD student from the University’s Institute of Integrative Biology, who worked on the project, said: “This high-resolution imaging technique allows us to study, for the first time, the assembly pattern of proteins in the shell architecture.
“It also provides the first clue about the motional dynamics of shell proteins in the formation of shell sheets. We can watch now how protein hexagons arrange in a honeycomb pattern in the shell. Such dynamics is required for the construction of the shell and may allow mature compartments to repair individual sides.”
In collaboration with scientists at the Lawrence Berkeley National Laboratory, the team used AFM and X-ray crystallography to further identify key protein residues that govern the interactions between shell proteins and the shell formation. This provides new clues to control the construction of the shell in artificial processes.
Pressing need
“Given their self-assembly nature and functional diversity, bacterial organelles constitute a rich source of metabolic modules for applications in synthetic biology. There is a pressing need to better understand how these compartments are formed from scratch by individual building blocks,” said Dr Luning Liu, a Royal Society University Research Fellow who led the study.
“Based on the pipeline developed, we can now seek to understand how a complete 3D structure of the compartment is built and how metabolites are sequestered within and transported across the shell.
“This work opens up a whole new way of exploring nature’s inner-cell architecture and functionality, which may aid researchers who are interested in engineering novel compartments or new types of nanoscale cages, such as ‘nanoreactors’ and drug-delivery systems. These will have impact on food and health security.”
The project was funded by the Biotechnology and Biological Sciences Research Council and the Royal Society.
“Visualization of bacterial microcompartment facet assembly using high-speed atomic force microscopy” is published as a cover article in Nano Letters (DOI: 10.1021/acs.nanolett.5b04259).
Great job!